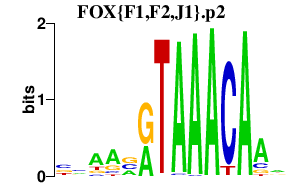
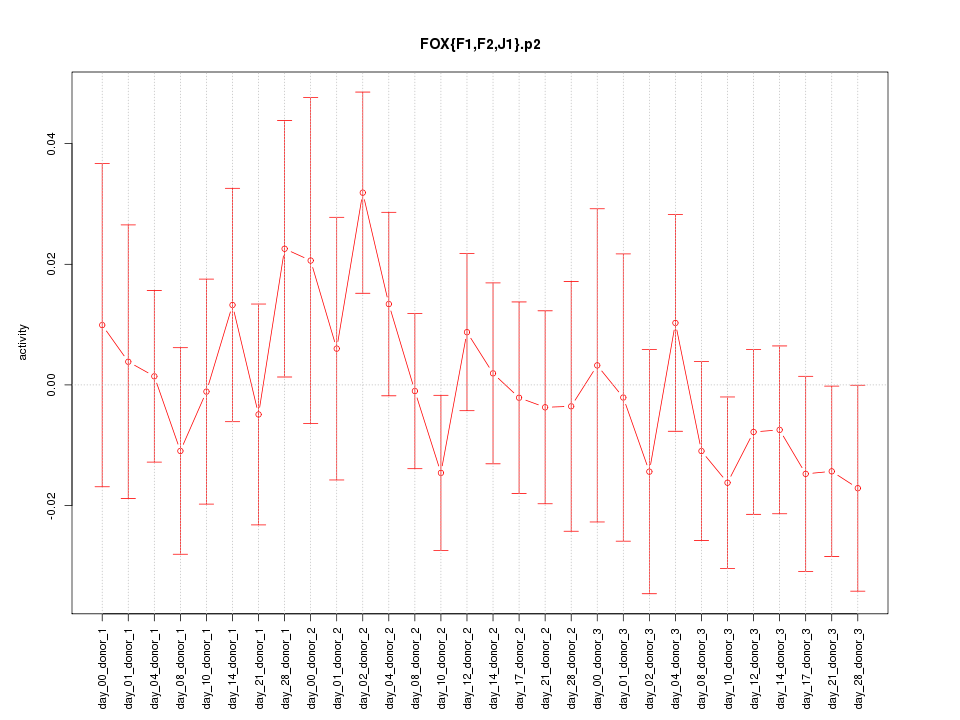
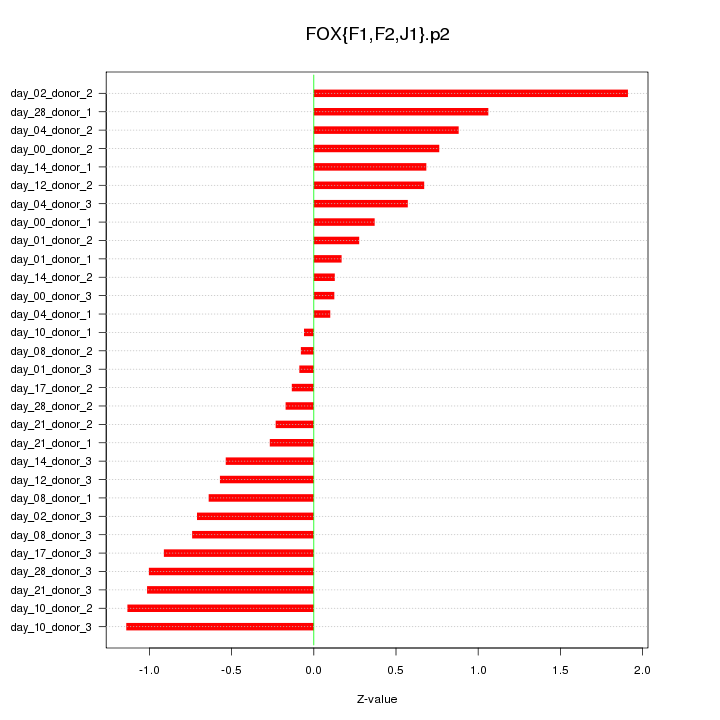
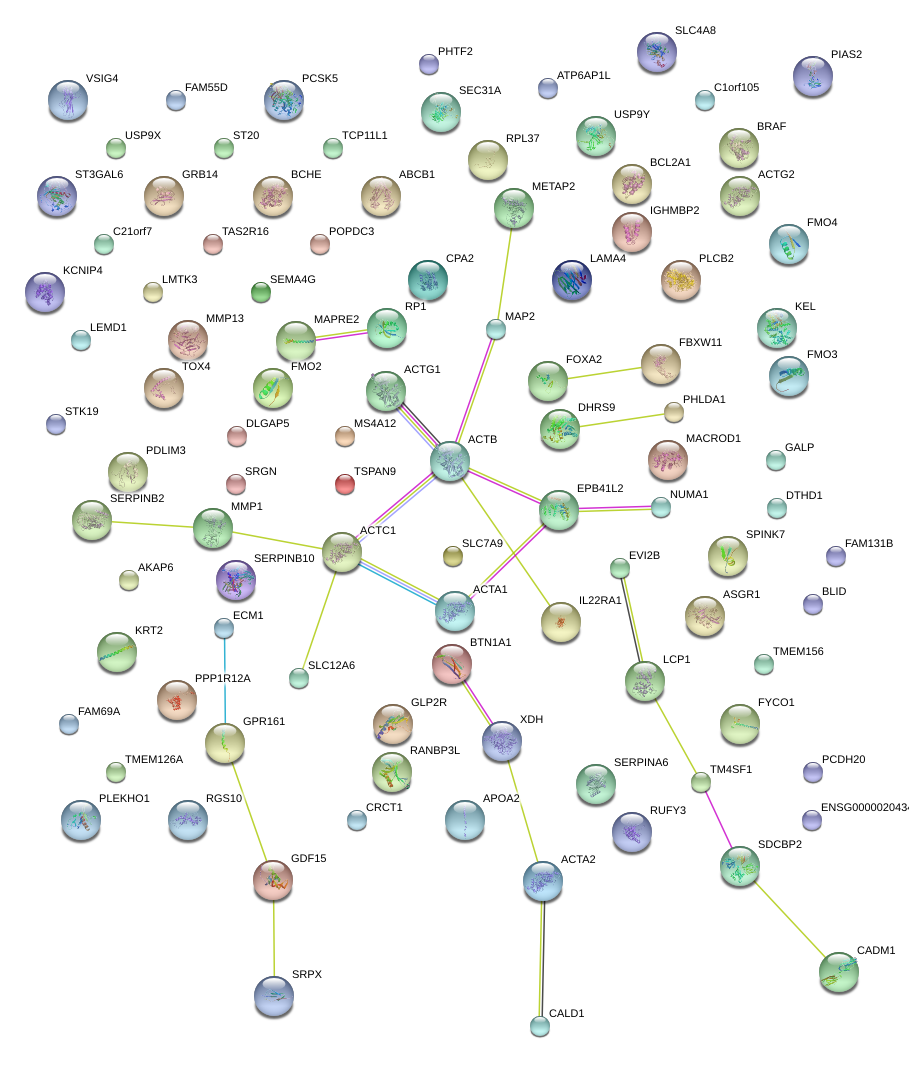
|
chr17_+_9745847
|
2.991
|
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr11_-_102826433
|
1.237
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chr11_-_102668892
|
1.230
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr11_-_102668822
|
1.190
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr6_-_131321868
|
1.176
|
NM_001252660
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr1_+_152486977
|
1.007
|
NM_019060
|
CRCT1
|
cysteine-rich C-terminal 1
|
|
chr2_-_165424993
|
0.926
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr1_-_24469571
|
0.867
|
NM_021258
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr1_+_171154387
|
0.823
|
NM_001460
|
FMO2
|
flavin containing monooxygenase 2 (non-functional)
|
|
chr11_-_121986800
|
0.769
|
NM_001001786
|
BLID
|
BH3-like motif containing, cell death inducer
|
|
chr7_-_122635753
|
0.742
|
NM_016945
|
TAS2R16
|
taste receptor, type 2, member 16
|
|
chr14_-_55650348
|
0.707
|
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr7_+_134576187
|
0.692
|
|
CALD1
|
caldesmon 1
|
|
chr11_-_71752099
|
0.681
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr7_+_77469446
|
0.677
|
NM_001127357
NM_001127359
NM_020432
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr4_-_39033981
|
0.671
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chrX_-_65259839
|
0.623
|
NM_001100431
NM_001184830
NM_001184831
NM_007268
|
VSIG4
|
V-set and immunoglobulin domain containing 4
|
|
chr1_-_93342706
|
0.613
|
NM_001252271
|
FAM69A
|
family with sequence similarity 69, member A
|
|
chr3_+_98451537
|
0.582
|
NM_006100
|
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6
|
|
chr4_-_186456580
|
0.571
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr4_-_21950373
|
0.564
|
NM_147182
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr17_-_5138049
|
0.550
|
NM_207103
|
SCIMP
|
SLP adaptor and CSK interacting membrane protein
|
|
chr5_+_147691988
|
0.549
|
NM_032566
|
SPINK7
|
serine peptidase inhibitor, Kazal type 7 (putative)
|
|
chr11_-_63933204
|
0.548
|
|
MACROD1
|
MACRO domain containing 1
|
|
chr1_+_150122415
|
0.548
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr1_-_205391180
|
0.528
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr14_+_32798478
|
0.526
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr17_-_29641068
|
0.526
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr5_+_81601165
|
0.525
|
NM_001017971
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like
|
|
chr7_+_129906702
|
0.520
|
NM_001869
|
CPA2
|
carboxypeptidase A2 (pancreatic)
|
|
chr8_+_55528626
|
0.519
|
NM_006269
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant)
|
|
chr4_+_36285643
|
0.518
|
NM_001170700
|
DTHD1
|
death domain containing 1
|
|
chr1_-_161193375
|
0.517
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr1_+_171060017
|
0.504
|
NM_001002294
NM_006894
|
FMO3
|
flavin containing monooxygenase 3
|
|
chr15_-_80207597
|
0.485
|
NM_001100879
|
ST20
|
suppressor of tumorigenicity 20
|
|
chr7_-_143059144
|
0.481
|
NM_014690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr19_+_56687388
|
0.475
|
NM_001145546
NM_033106
|
GALP
|
galanin-like peptide
|
|
chr10_-_121296044
|
0.459
|
NM_002925
|
RGS10
|
regulator of G-protein signaling 10
|
|
chr2_+_210444154
|
0.456
|
|
MAP2
|
microtubule-associated protein 2
|
|
chrY_+_14813159
|
0.450
|
NM_004654
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked
|
|
chr9_+_78505514
|
0.449
|
NM_001190482
NM_006200
|
PCSK5
|
proprotein convertase subtilisin/kexin type 5
|
|
chr3_-_165555252
|
0.445
|
NM_000055
|
BCHE
|
butyrylcholinesterase
|
|
chr17_-_7082559
|
0.441
|
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr20_-_1309809
|
0.436
|
NM_080489
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr21_+_30517897
|
0.430
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr11_-_115375065
|
0.430
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr7_-_142659397
|
0.426
|
|
KEL
|
Kell blood group, metallo-endopeptidase
|
|
chr15_-_34610928
|
0.423
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr12_-_80307690
|
0.416
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr13_-_46756291
|
0.415
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr1_+_209602164
|
0.410
|
NM_001104548
|
MIR205HG
|
MIR205 host gene (non-protein coding)
|
|
chr12_-_76425375
|
0.407
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr3_-_165555167
|
0.406
|
|
BCHE
|
butyrylcholinesterase
|
|
chr1_+_150480486
|
0.405
|
NM_001202858
NM_004425
NM_022664
|
ECM1
|
extracellular matrix protein 1
|
|
chr17_-_7082654
|
0.400
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr19_-_49016418
|
0.399
|
NM_001080434
|
LMTK3
|
lemur tyrosine kinase 3
|
|
chr11_-_63933493
|
0.388
|
NM_014067
|
MACROD1
|
MACRO domain containing 1
|
|
chr20_-_22566092
|
0.381
|
NM_153675
|
FOXA2
|
forkhead box A2
|
|
chr1_-_168106810
|
0.381
|
|
GPR161
|
G protein-coupled receptor 161
|
|
chr11_+_60260250
|
0.372
|
NM_001164470
NM_017716
|
MS4A12
|
membrane-spanning 4-domains, subfamily A, member 12
|
|
chr4_+_71587674
|
0.370
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr18_+_61575203
|
0.368
|
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr19_+_18391252
|
0.368
|
|
|
|
|
chr10_-_90712510
|
0.365
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr2_-_31637553
|
0.364
|
|
XDH
|
xanthine dehydrogenase
|
|
chr11_-_67450536
|
0.363
|
|
RPL37
|
ribosomal protein L37
|
|
chrX_-_38079980
|
0.363
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr1_-_168106727
|
0.358
|
|
GPR161
|
G protein-coupled receptor 161
|
|
chr10_+_102732285
|
0.353
|
NM_001203244
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr10_+_22540986
|
0.350
|
|
LOC100130992
|
uncharacterized LOC100130992
|
|
chr2_+_169929009
|
0.349
|
NM_001142270
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr17_-_29641094
|
0.344
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr11_-_114466471
|
0.344
|
NM_001077639
NM_017678
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr1_+_150122095
|
0.343
|
NM_016274
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr12_-_53045947
|
0.334
|
NM_000423
|
KRT2
|
keratin 2
|
|
chr16_-_86542463
|
0.332
|
|
LOC400550
|
uncharacterized LOC400550
|
|
chr6_-_105627857
|
0.331
|
NM_022361
|
POPDC3
|
popeye domain containing 3
|
|
chr3_-_149086884
|
0.329
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr11_+_33060962
|
0.328
|
NM_018393
|
TCP11L1
|
t-complex 11 (mouse)-like 1
|
|
chr19_-_33360641
|
0.327
|
NM_001126335
NM_001243036
NM_014270
|
SLC7A9
|
solute carrier family 7 (glycoprotein-associated amino acid transporter light chain, bo,+ system), member 9
|
|
chr7_-_140624280
|
0.325
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr1_+_150122246
|
0.323
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr19_+_18496966
|
0.320
|
NM_004864
|
GDF15
|
growth differentiation factor 15
|
|
chr14_-_94789641
|
0.320
|
NM_001756
|
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6
|
|
chr5_-_36301999
|
0.318
|
NM_001161429
NM_145000
|
RANBP3L
|
RAN binding protein 3-like
|
|
chr15_-_40600065
|
0.317
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr10_+_70847817
|
0.315
|
NM_002727
|
SRGN
|
serglycin
|
|
chr1_+_172389827
|
0.315
|
NM_139240
|
C1orf105
|
chromosome 1 open reading frame 105
|
|
chr6_+_26501448
|
0.314
|
NM_001732
|
BTN1A1
|
butyrophilin, subfamily 1, member A1
|
|
chr6_-_112575766
|
0.312
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr1_-_168106535
|
0.302
|
|
GPR161
|
G protein-coupled receptor 161
|
|
chr7_-_87342569
|
0.299
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr15_-_40599974
|
0.298
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr11_+_33061560
|
0.295
|
NM_001145541
|
TCP11L1
|
t-complex 11 (mouse)-like 1
|
|
chr13_-_61989209
|
0.295
|
|
PCDH20
|
protocadherin 20
|
|
chr6_-_112575880
|
0.295
|
|
LAMA4
|
laminin, alpha 4
|
|
chr22_-_33402648
|
0.293
|
NM_003490
NM_133633
|
SYN3
|
synapsin III
|
|
chr4_-_89978117
|
0.293
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr9_-_21305311
|
0.290
|
NM_002169
|
IFNA5
|
interferon, alpha 5
|
|
chr12_-_99038705
|
0.289
|
|
IKBIP
|
IKBKB interacting protein
|
|
chr8_-_72459891
|
0.288
|
|
|
|
|
chr7_-_142659412
|
0.288
|
NM_000420
|
KEL
|
Kell blood group, metallo-endopeptidase
|
|
chr11_-_27723061
|
0.283
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr17_-_29641087
|
0.283
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr6_-_112575708
|
0.282
|
|
LAMA4
|
laminin, alpha 4
|
|
chr14_+_74551655
|
0.282
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr8_+_1952875
|
0.282
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr13_+_98086315
|
0.282
|
NM_021033
|
RAP2A
|
RAP2A, member of RAS oncogene family
|
|
chr2_+_33661392
|
0.281
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr8_-_15095791
|
0.281
|
NM_139167
|
SGCZ
|
sarcoglycan, zeta
|
|
chr3_-_71180061
|
0.279
|
NM_001244815
|
FOXP1
|
forkhead box P1
|
|
chr7_+_123249076
|
0.278
|
NM_080928
|
ASB15
|
ankyrin repeat and SOCS box containing 15
|
|
chr7_+_128312345
|
0.276
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr11_-_59612925
|
0.274
|
NM_005142
|
GIF
|
gastric intrinsic factor (vitamin B synthesis)
|
|
chr10_+_97759847
|
0.273
|
NM_001001732
NM_001159747
|
CC2D2B
|
coiled-coil and C2 domain containing 2B
|
|
chr20_+_44036940
|
0.273
|
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr18_+_60382617
|
0.271
|
NM_194449
|
PHLPP1
|
PH domain and leucine rich repeat protein phosphatase 1
|
|
chr3_-_167191808
|
0.267
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr6_-_30655671
|
0.267
|
NM_133471
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr5_-_39203061
|
0.266
|
|
FYB
|
FYN binding protein
|
|
chr15_+_80390757
|
0.265
|
NM_001242919
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr4_-_152149042
|
0.263
|
NM_001243349
|
SH3D19
|
SH3 domain containing 19
|
|
chr6_-_128239741
|
0.262
|
NM_001164687
|
THEMIS
|
thymocyte selection associated
|
|
chr7_+_134233848
|
0.260
|
NM_001080538
|
AKR1B15
|
aldo-keto reductase family 1, member B15
|
|
chr5_-_132161863
|
0.259
|
NM_133456
|
SHROOM1
|
shroom family member 1
|
|
chr13_-_99910548
|
0.258
|
NM_001098200
NM_005292
|
GPR18
|
G protein-coupled receptor 18
|
|
chr22_+_37309618
|
0.258
|
NM_000395
|
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage)
|
|
chr1_-_161193190
|
0.253
|
|
APOA2
|
apolipoprotein A-II
|
|
chr5_-_180076599
|
0.253
|
|
FLT4
|
fms-related tyrosine kinase 4
|
|
chr1_-_197036350
|
0.252
|
NM_001994
|
F13B
|
coagulation factor XIII, B polypeptide
|
|
chr15_-_40600025
|
0.250
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr15_+_43477315
|
0.250
|
NM_012142
|
CCNDBP1
|
cyclin D-type binding-protein 1
|
|
chr19_-_46148606
|
0.249
|
NM_001193268
|
EML2
|
echinoderm microtubule associated protein like 2
|
|
chr3_-_20053618
|
0.248
|
NM_001252657
|
PP2D1
|
protein phosphatase 2C-like domain containing 1
|
|
chr1_+_84630374
|
0.245
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr10_-_5045967
|
0.243
|
NM_001135241
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr8_+_74903563
|
0.242
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr8_+_120079423
|
0.241
|
NM_006438
|
COLEC10
|
collectin sub-family member 10 (C-type lectin)
|
|
chr1_-_186649421
|
0.241
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr4_+_155484131
|
0.236
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr18_+_29769986
|
0.235
|
NM_005925
|
MEP1B
|
meprin A, beta
|
|
chr3_-_71179715
|
0.234
|
|
FOXP1
|
forkhead box P1
|
|
chr2_-_192711651
|
0.233
|
|
SDPR
|
serum deprivation response
|
|
chr4_-_123377471
|
0.233
|
NM_000586
|
IL2
|
interleukin 2
|
|
chr2_-_165478357
|
0.232
|
NM_004490
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr12_-_96390002
|
0.232
|
NM_002108
|
HAL
|
histidine ammonia-lyase
|
|
chr3_+_156392204
|
0.231
|
NM_015508
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr1_+_192544856
|
0.230
|
NM_002922
|
RGS1
|
regulator of G-protein signaling 1
|
|
chr4_-_111119752
|
0.228
|
NM_001130721
NM_024090
|
ELOVL6
|
ELOVL fatty acid elongase 6
|
|
chr9_-_21368005
|
0.228
|
NM_006900
|
IFNA1
IFNA13
|
interferon, alpha 1
interferon, alpha 13
|
|
chr18_+_61370193
|
0.227
|
NM_080475
|
SERPINB11
|
serpin peptidase inhibitor, clade B (ovalbumin), member 11 (gene/pseudogene)
|
|
chr3_+_143692166
|
0.226
|
NM_001134470
|
C3orf58
|
chromosome 3 open reading frame 58
|
|
chr16_+_56225301
|
0.226
|
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr5_-_13944588
|
0.226
|
NM_001369
|
DNAH5
|
dynein, axonemal, heavy chain 5
|
|
chr2_+_179184970
|
0.225
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr1_+_74701070
|
0.224
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr4_-_70826712
|
0.224
|
NM_001891
|
CSN2
|
casein beta
|
|
chr3_-_150920946
|
0.221
|
NM_013308
|
GPR171
|
G protein-coupled receptor 171
|
|
chr7_+_37723378
|
0.220
|
|
|
|
|
chr9_+_136287119
|
0.220
|
NM_139025
NM_139026
NM_139027
|
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13
|
|
chr1_+_20915440
|
0.220
|
NM_001785
|
CDA
|
cytidine deaminase
|
|
chr1_+_169337432
|
0.219
|
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr16_+_81040120
|
0.217
|
|
CENPN
|
centromere protein N
|
|
chr11_-_5526833
|
0.212
|
NM_001005567
|
OR51B5
HBE1
|
olfactory receptor, family 51, subfamily B, member 5
hemoglobin, epsilon 1
|
|
chr1_-_103496768
|
0.211
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr14_+_37126772
|
0.210
|
NM_006194
|
PAX9
|
paired box 9
|
|
chr5_-_10308167
|
0.209
|
NM_138809
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas)
|
|
chr15_+_40509628
|
0.209
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr8_-_108510094
|
0.209
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr1_+_116654375
|
0.208
|
NM_152367
|
MAB21L3
|
mab-21-like 3 (C. elegans)
|
|
chr1_-_240775388
|
0.208
|
|
GREM2
|
gremlin 2
|
|
chr6_+_108882104
|
0.207
|
|
FOXO3
|
forkhead box O3
|
|
chr20_+_16728997
|
0.206
|
NM_020157
|
OTOR
|
otoraplin
|
|
chr14_+_65877309
|
0.205
|
NM_004480
NM_178155
NM_178156
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase)
|
|
chr13_+_41885340
|
0.204
|
NM_001110798
NM_018527
NM_024561
|
NAA16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit
|
|
chr1_-_169703178
|
0.204
|
NM_000450
|
SELE
|
selectin E
|
|
chr7_+_28725597
|
0.203
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr16_+_28699878
|
0.203
|
NM_001199142
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr3_+_156392369
|
0.202
|
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr12_+_9142136
|
0.196
|
NM_005810
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1
|
|
chr2_-_192711923
|
0.195
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr19_-_51141301
|
0.194
|
NM_032298
|
SYT3
|
synaptotagmin III
|
|
chr1_+_203444955
|
0.194
|
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr1_-_231560789
|
0.194
|
NM_022051
|
EGLN1
|
egl nine homolog 1 (C. elegans)
|
|
chr2_-_215896809
|
0.193
|
NM_015657
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr1_+_65730423
|
0.193
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr5_-_36241684
|
0.193
|
|
NADKD1
|
NAD kinase domain containing 1
|
|
chr17_+_58755158
|
0.192
|
NM_001099432
NM_017679
|
BCAS3
|
breast carcinoma amplified sequence 3
|
|
chr12_+_79258588
|
0.191
|
|
SYT1
|
synaptotagmin I
|
|
chr9_+_136287443
|
0.189
|
|
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13
|
|
chr5_+_60933635
|
0.188
|
NM_173667
|
C5orf64
|
chromosome 5 open reading frame 64
|
|
chr12_+_100041527
|
0.188
|
NM_153364
|
FAM71C
|
family with sequence similarity 71, member C
|
|
chr10_+_70847857
|
0.188
|
|
SRGN
|
serglycin
|
|
chr13_+_109248499
|
0.187
|
NM_015011
|
MYO16
|
myosin XVI
|
|
chr6_-_127780534
|
0.186
|
NM_014702
|
KIAA0408
|
KIAA0408
|
|
chr13_-_45775174
|
0.185
|
NM_198404
|
KCTD4
|
potassium channel tetramerisation domain containing 4
|
|
chr22_+_50609159
|
0.185
|
NM_001160300
NM_052839
|
PANX2
|
pannexin 2
|
|
chr6_-_76203256
|
0.185
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chrX_-_15288172
|
0.185
|
NM_001031739
NM_001168530
NM_024087
|
ASB9
|
ankyrin repeat and SOCS box containing 9
|
|
chr9_-_77503009
|
0.184
|
NM_017662
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6
|